Exploring the chemical space by Principal Component Analysis (PCA) and clustering
In this chem-workflow, I will show you how to analyze compound databases in order to identify shared descriptors (physicochemical properties) among them.
A server to perform this analysis (or a similar one) is Platform for Unified Molecular Analysis (PUMA).
For this purpose, I will use a fraction of the REAL Compound Library of Enamine. You can find another example using the same database here.
Importing the libraries we will use
[1]:
import os
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import gridspec
from rdkit import Chem, DataStructs
from rdkit.Chem import Descriptors,Crippen
from sklearn.decomposition import PCA
from sklearn.preprocessing import StandardScaler
import matplotlib.cm as cm
from sklearn.metrics import silhouette_samples, silhouette_score
from sklearn.cluster import KMeans
Loading the molecules
The library contains more than 8 000 000 SMILES codes (size 542.8 M). But we can read the .smiles file as a text file to keep the fist 100 molecules in a pandas table.
[2]:
# The result of this code will be:
table=pd.DataFrame()
with open('Enamine_REAL_diversity.smiles','r') as file:
for index,line in enumerate(file):
if 0<index<=100:
table.loc[index,'SMILES']=line.split()[0]
table.loc[index,'Id']=line.split()[1]
Let´s take a look at the first 10 elements of our table of compounds.
[3]:
table.head(10)
[3]:
| SMILES | Id | |
|---|---|---|
| 1 | CC(C)C(C)(C)SCC=CBr | PV-002312932038 |
| 2 | CC1CC2(NC(C3CC3C)=NO2)C(C)O1 | Z3092470692 |
| 3 | BrC=CCN1CC2COC(C2)C1 | Z3310927277 |
| 4 | CC(C)CC(C)C(C)NCC1(C)CSC1 | Z3362561558 |
| 5 | CC(C)(C)CS(=O)CC=CCl | Z2904173473 |
| 6 | CC(C)(C)C1=NOC2(N1)[C@H]1OC[C@H]3C[C@@H]2C[C@@... | Z3092410916 |
| 7 | BrC=CCNC1CCCC=C1Br | Z3322438014 |
| 8 | CC(C)C1CC(NCCC2SCCS2)CN1 | Z3350500536 |
| 9 | FCC(CF)NCC1(C(F)F)CC1 | PV-002106209714 |
| 10 | CC(C(F)F)N1N=C(C(F)(F)F)C(F)C1=O | Z2790107381 |
Molecular descriptors calculation
We can use RDKIT to calculate several molecular descriptors (2D and 3D). However, for this example, we will focus on the descriptors measured in the publication: Platform for Unified Molecular Analysis PUMA 10.1021/acs.jcim.7b00253.
Moreover, a list of all descriptor that can be calculated using RDKIT can be found here.
[4]:
# We will calculate the descriptors and add them to our table
for i in table.index:
mol=Chem.MolFromSmiles(table.loc[i,'SMILES'])
table.loc[i,'MolWt']=Descriptors.ExactMolWt (mol)
table.loc[i,'TPSA']=Chem.rdMolDescriptors.CalcTPSA(mol) #Topological Polar Surface Area
table.loc[i,'nRotB']=Descriptors.NumRotatableBonds (mol) #Number of rotable bonds
table.loc[i,'HBD']=Descriptors.NumHDonors(mol) #Number of H bond donors
table.loc[i,'HBA']=Descriptors.NumHAcceptors(mol) #Number of H bond acceptors
table.loc[i,'LogP']=Descriptors.MolLogP(mol) #LogP
As a result, we will get a table with all descriptors for each molecule (SMILES code).
[5]:
table.head(10)
[5]:
| SMILES | Id | MolWt | TPSA | nRotB | HBD | HBA | LogP | |
|---|---|---|---|---|---|---|---|---|
| 1 | CC(C)C(C)(C)SCC=CBr | PV-002312932038 | 236.023434 | 0.00 | 4.0 | 0.0 | 1.0 | 4.0628 |
| 2 | CC1CC2(NC(C3CC3C)=NO2)C(C)O1 | Z3092470692 | 210.136828 | 42.85 | 1.0 | 1.0 | 4.0 | 1.4693 |
| 3 | BrC=CCN1CC2COC(C2)C1 | Z3310927277 | 231.025876 | 12.47 | 2.0 | 0.0 | 2.0 | 1.6157 |
| 4 | CC(C)CC(C)C(C)NCC1(C)CSC1 | Z3362561558 | 229.186421 | 12.03 | 6.0 | 1.0 | 2.0 | 3.3998 |
| 5 | CC(C)(C)CS(=O)CC=CCl | Z2904173473 | 194.053214 | 17.07 | 3.0 | 0.0 | 1.0 | 2.5337 |
| 6 | CC(C)(C)C1=NOC2(N1)[C@H]1OC[C@H]3C[C@@H]2C[C@@... | Z3092410916 | 236.152478 | 42.85 | 0.0 | 1.0 | 4.0 | 1.7169 |
| 7 | BrC=CCNC1CCCC=C1Br | Z3322438014 | 292.941474 | 12.03 | 3.0 | 1.0 | 1.0 | 3.3159 |
| 8 | CC(C)C1CC(NCCC2SCCS2)CN1 | Z3350500536 | 260.138091 | 24.06 | 5.0 | 2.0 | 4.0 | 2.1587 |
| 9 | FCC(CF)NCC1(C(F)F)CC1 | PV-002106209714 | 199.098412 | 12.03 | 6.0 | 1.0 | 1.0 | 1.9289 |
| 10 | CC(C(F)F)N1N=C(C(F)(F)F)C(F)C1=O | Z2790107381 | 248.038432 | 32.67 | 2.0 | 0.0 | 2.0 | 1.7386 |
Principal Component Analysis of calculated molecular descriptors (PCA)
I won´t discuss the basis of PCA due to this site is focused on workflows. However, PCA analysis for different application has been discussed plenty. As a quick reference, you can visit the PCA explanation at Wikipedia.
For our analysis, we need to select the values of the table that we will use for PCA. Meaning, the descriptors we calculated.
[6]:
descriptors = table.loc[:, ['MolWt', 'TPSA', 'nRotB', 'HBD','HBA', 'LogP']].values
A very important step is performing a standardization of the scales of the descriptors. Scales differences in PCA modify the variance distribution during PCA. More info about this topic can be found here.
[7]:
descriptors_std = StandardScaler().fit_transform(descriptors)
Now, we are ready to calculate the PCA
[8]:
pca = PCA()
descriptors_2d = pca.fit_transform(descriptors_std)
Let´s add the PCA data to a new table
[9]:
descriptors_pca= pd.DataFrame(descriptors_2d)
descriptors_pca.index = table.index
descriptors_pca.columns = ['PC{}'.format(i+1) for i in descriptors_pca.columns]
descriptors_pca.head(10)
[9]:
| PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | |
|---|---|---|---|---|---|---|
| 1 | -2.463784 | -0.936078 | -0.274030 | 0.558761 | -0.063431 | 0.131835 |
| 2 | -0.075200 | 1.908703 | -0.534952 | 0.042434 | -0.559363 | -0.252494 |
| 3 | -1.477081 | 0.865532 | -1.070530 | -0.451302 | 0.453224 | -0.201938 |
| 4 | -1.526756 | -0.336039 | 1.094536 | 0.286876 | -0.170061 | -0.175008 |
| 5 | -2.147808 | 0.475692 | -0.564722 | -0.344079 | -0.139551 | 0.425832 |
| 6 | 0.066967 | 1.802380 | -0.849193 | 0.543555 | -0.370788 | -0.232141 |
| 7 | -1.384515 | -0.041261 | 0.202822 | 1.299544 | 0.823427 | 0.123817 |
| 8 | 0.083638 | 0.888434 | 1.430529 | 0.522356 | 0.088537 | -0.931776 |
| 9 | -1.817628 | 0.741035 | 1.132660 | -0.646553 | 0.332754 | -0.019503 |
| 10 | -0.899886 | 0.776893 | -1.091563 | -0.339891 | 0.401541 | 0.382843 |
We can check the explained variance to see the variance explained by each component from PCA
[10]:
print(pca.explained_variance_ratio_)
print(sum(pca.explained_variance_ratio_))
[0.42278681 0.30201227 0.14303101 0.08478587 0.03041105 0.01697299]
1.0
And also, we can plot such data
[11]:
plt.rcParams['axes.linewidth'] = 1.5
plt.figure(figsize=(8,6))
fig, ax = plt.subplots(figsize=(8,6))
var=np.cumsum(np.round(pca.explained_variance_ratio_, decimals=3)*100)
plt.plot([i+1 for i in range(len(var))],var,'k-',linewidth=2)
plt.xticks([i+1 for i in range(len(var))])
plt.ylabel('% Variance Explained',fontsize=16,fontweight='bold')
plt.xlabel('Pincipal Component (PC)',fontsize=16,fontweight='bold')
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
plt.tight_layout()
plt.tick_params ('both',width=2,labelsize=12)
<Figure size 576x432 with 0 Axes>
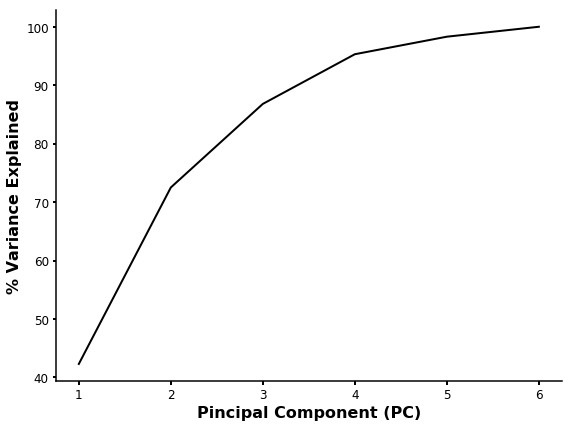
As you can see, the PC1 and PC2 explain 72.4 % of the variability. So we can plot PC1 vs PC2 to see the distribution of our compounds.
[12]:
fig = plt.figure(figsize=(8,6))
ax = fig.add_subplot(111)
ax.plot(descriptors_pca['PC1'],descriptors_pca['PC2'],'o',color='k')
ax.set_title ('Principal Component Analysis',fontsize=16,fontweight='bold',family='sans-serif')
ax.set_xlabel ('PC1',fontsize=14,fontweight='bold')
ax.set_ylabel ('PC2',fontsize=14,fontweight='bold')
plt.tick_params ('both',width=2,labelsize=12)
plt.tight_layout()
plt.show()
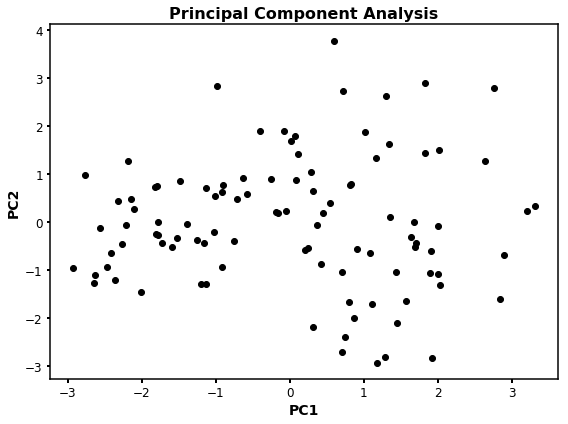
However, this plot is simple and we cannot identify compound clusters easily. For such purpose, we can perform clustering analysis using the PCA values to identify compound groups by mathematical approaches. Moreover, PC1 vs PC2 (or any other combination) won´t give us information about which feature (descriptor) is more important to explain the variance of our values.
For this example, we will identify the most important feature (descriptor), and we identify compound clusters by the k-means clustering algorithm. For more info about k-means, you can look at skit-learn.
K-means clustering and main features identification
The first step for this analysis is to re-scale our PCA values from -1 to 1. This because we want to analyze our data inside of the covariance cycle of the features (descriptors). Fur such purpose we type:
[13]:
# This normalization will be performed just for PC1 and PC2, but can be done for all the components.
scale1 = 1.0/(max(descriptors_pca['PC1']) - min(descriptors_pca['PC1']))
scale2 = 1.0/(max(descriptors_pca['PC2']) - min(descriptors_pca['PC2']))
# And we add the new values to our PCA table
descriptors_pca['PC1_normalized']=[i*scale1 for i in descriptors_pca['PC1']]
descriptors_pca['PC2_normalized']=[i*scale2 for i in descriptors_pca['PC2']]
[14]:
descriptors_pca.head(10)
[14]:
| PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC1_normalized | PC2_normalized | |
|---|---|---|---|---|---|---|---|---|
| 1 | -2.463784 | -0.936078 | -0.274030 | 0.558761 | -0.063431 | 0.131835 | -0.395072 | -0.139342 |
| 2 | -0.075200 | 1.908703 | -0.534952 | 0.042434 | -0.559363 | -0.252494 | -0.012058 | 0.284125 |
| 3 | -1.477081 | 0.865532 | -1.070530 | -0.451302 | 0.453224 | -0.201938 | -0.236852 | 0.128841 |
| 4 | -1.526756 | -0.336039 | 1.094536 | 0.286876 | -0.170061 | -0.175008 | -0.244818 | -0.050022 |
| 5 | -2.147808 | 0.475692 | -0.564722 | -0.344079 | -0.139551 | 0.425832 | -0.344405 | 0.070810 |
| 6 | 0.066967 | 1.802380 | -0.849193 | 0.543555 | -0.370788 | -0.232141 | 0.010738 | 0.268298 |
| 7 | -1.384515 | -0.041261 | 0.202822 | 1.299544 | 0.823427 | 0.123817 | -0.222009 | -0.006142 |
| 8 | 0.083638 | 0.888434 | 1.430529 | 0.522356 | 0.088537 | -0.931776 | 0.013412 | 0.132250 |
| 9 | -1.817628 | 0.741035 | 1.132660 | -0.646553 | 0.332754 | -0.019503 | -0.291460 | 0.110309 |
| 10 | -0.899886 | 0.776893 | -1.091563 | -0.339891 | 0.401541 | 0.382843 | -0.144298 | 0.115646 |
[15]:
fig = plt.figure(figsize=(8,6))
ax = fig.add_subplot(111)
ax.plot(descriptors_pca['PC1_normalized'],descriptors_pca['PC2_normalized'],'o',color='k')
ax.set_title ('Principal Component Analysis',fontsize=16,fontweight='bold',family='sans-serif')
ax.set_xlabel ('PC1',fontsize=14,fontweight='bold')
ax.set_ylabel ('PC2',fontsize=14,fontweight='bold')
plt.tick_params ('both',width=2,labelsize=12)
plt.tight_layout()
plt.show()
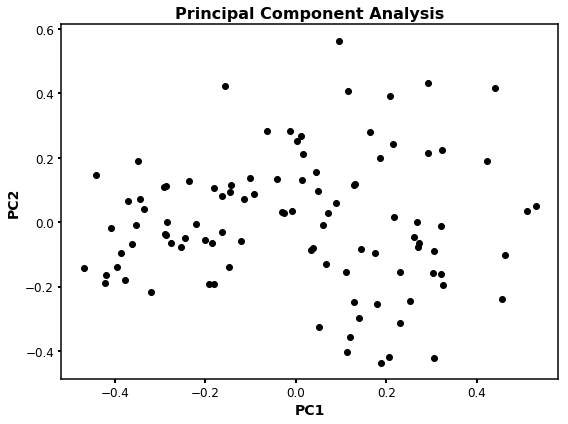
As you can see, the distribution of the points is the same as before, however, the scale now is from -1 to 1.
k-means clustering
K-means clustering is an algorithm in which the user must define the number of clusters. However, in order to mathematically select a number of clusters for a group of points based on distribution, different algorithms can be applied. For instance, we will use the silhouette-based algorithm to identify the best number of clusters for our distribution. More info about silhouette algorithm can be found here and here.
[16]:
range_n_clusters = [2, 3, 4, 5, 6, 7,8,9,10]
for n_clusters in range_n_clusters:
fig, (ax1,ax2)= plt.subplots(1, 2)
fig.set_size_inches(8, 4)
kmeans = KMeans(n_clusters=n_clusters, random_state=10)
cluster_labels = kmeans.fit_predict(descriptors_pca[['PC1_normalized','PC2_normalized']])
silhouette_avg = silhouette_score(descriptors_pca[['PC1_normalized','PC2_normalized']], cluster_labels)
print("For n_clusters =", n_clusters,
"The average silhouette_score is :", silhouette_avg)
sample_silhouette_values = silhouette_samples(descriptors_pca[['PC1_normalized','PC2_normalized']], cluster_labels)
y_lower = 10
for i in range(n_clusters):
# Aggregate the silhouette scores for samples belonging to
# cluster i, and sort them
ith_cluster_silhouette_values = \
sample_silhouette_values[cluster_labels == i]
ith_cluster_silhouette_values.sort()
size_cluster_i = ith_cluster_silhouette_values.shape[0]
y_upper = y_lower + size_cluster_i
color = cm.nipy_spectral(float(i) / n_clusters)
ax1.fill_betweenx(np.arange(y_lower, y_upper),
0, ith_cluster_silhouette_values,
facecolor=color, edgecolor=color, alpha=0.7)
# Label the silhouette plots with their cluster numbers at the middle
ax1.text(-0.05, y_lower + 0.5 * size_cluster_i, str(i))
# Compute the new y_lower for next plot
y_lower = y_upper + 10 # 10 for the 0 samples
ax1.set_title("The silhouette plot for the various clusters.")
ax1.set_xlabel("The silhouette coefficient values")
ax1.set_ylabel("Cluster label")
# The vertical line for average silhouette score of all the values
ax1.axvline(x=silhouette_avg, color="red", linestyle="--")
# 2nd Plot showing the actual clusters formed
colors = cm.nipy_spectral(cluster_labels.astype(float) / n_clusters)
ax2.scatter(descriptors_pca['PC1_normalized'], descriptors_pca['PC2_normalized'],
marker='.', s=30, lw=0, alpha=0.7,c=colors, edgecolor='k')
# Labeling the clusters
centers = kmeans.cluster_centers_
# Draw white circles at cluster centers
ax2.scatter(centers[:, 0], centers[:, 1], marker='o',
c="white", alpha=1, s=200, edgecolor='k')
for i, c in enumerate(centers):
ax2.scatter(c[0], c[1], marker='$%d$' % i, alpha=1,
s=50, edgecolor='k')
ax2.set_title("The visualization of the clustered data.")
ax2.set_xlabel("PC1")
ax2.set_ylabel("PC2")
plt.suptitle(("Silhouette analysis for KMeans clustering on sample data "
"with n_clusters = %d" % n_clusters),
fontsize=14, fontweight='bold')
plt.show()
For n_clusters = 2 The average silhouette_score is : 0.4084228227138712
For n_clusters = 3 The average silhouette_score is : 0.45489833249431
For n_clusters = 4 The average silhouette_score is : 0.4241945808059022
For n_clusters = 5 The average silhouette_score is : 0.4131292685513735
For n_clusters = 6 The average silhouette_score is : 0.39907782063536723
For n_clusters = 7 The average silhouette_score is : 0.3523945319600729
For n_clusters = 8 The average silhouette_score is : 0.3906505698840795
For n_clusters = 9 The average silhouette_score is : 0.4003463850492715
For n_clusters = 10 The average silhouette_score is : 0.384264972688578
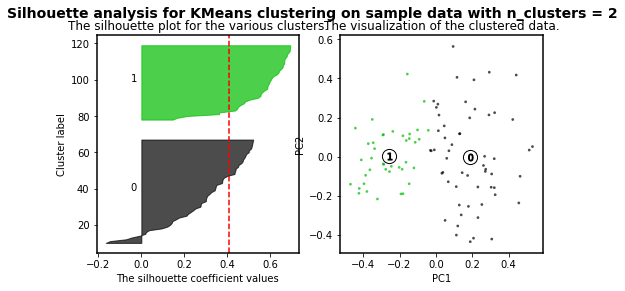
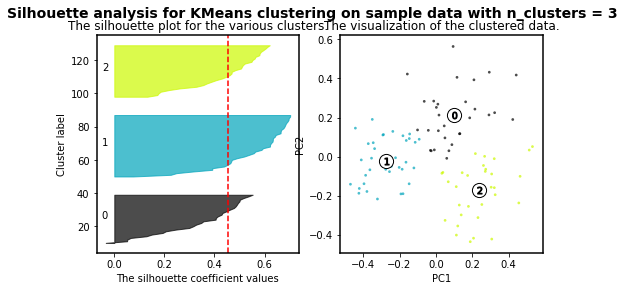
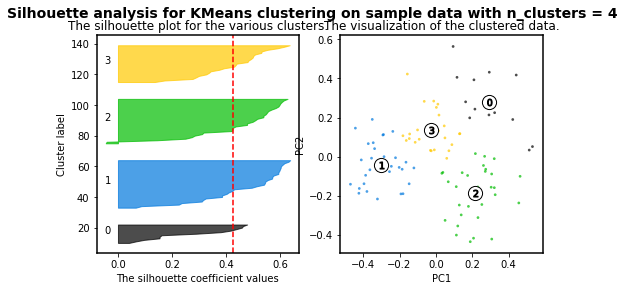
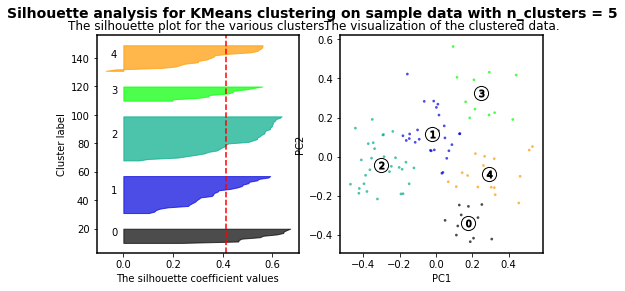
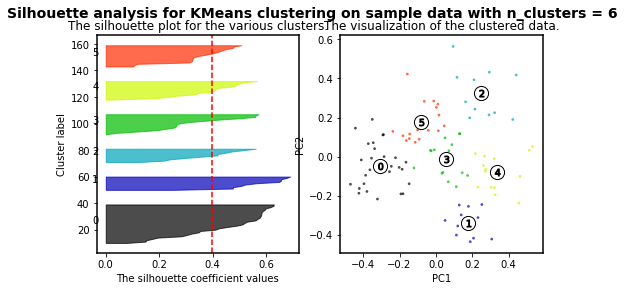
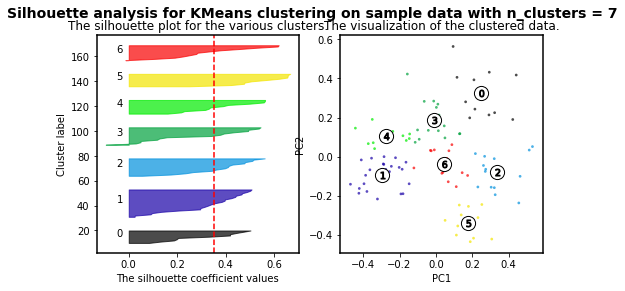
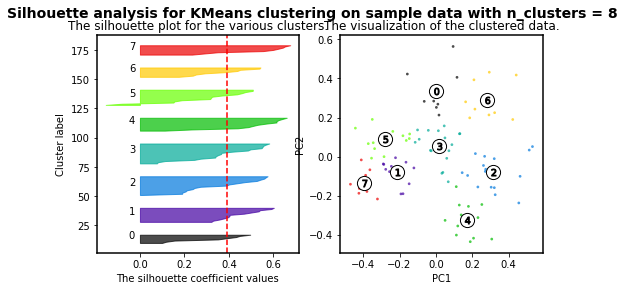
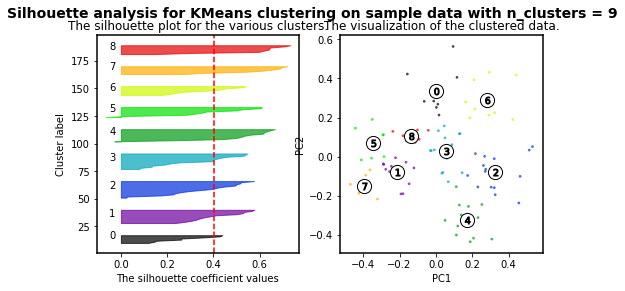
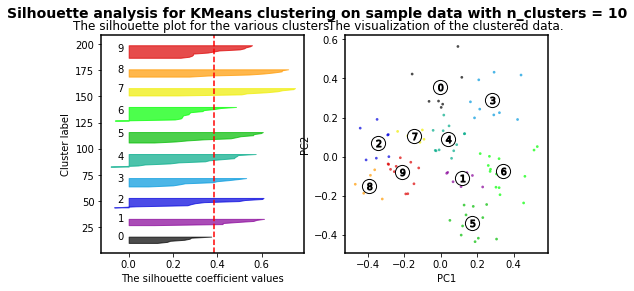
As higher the silhouette_score the better cluster distribution. Then, for our data:
For n_clusters = 3 The average silhouette_score is : 0.45489833249431
3 clusters are the best for our data distribution. So, let´s use such number of clusters.
[17]:
kmeans = KMeans(n_clusters=3, random_state=10) # We define the best number of clusters
clusters = kmeans.fit(descriptors_pca[['PC1_normalized','PC2_normalized']]) #PC1 vs PC2 (normalized values)
Once the calculation of clusters is done, we can add the result to our PCA table.
[18]:
descriptors_pca['Cluster_PC1_PC2'] = pd.Series(clusters.labels_, index=table.index)
[19]:
descriptors_pca.head(10)
[19]:
| PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC1_normalized | PC2_normalized | Cluster_PC1_PC2 | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | -2.463784 | -0.936078 | -0.274030 | 0.558761 | -0.063431 | 0.131835 | -0.395072 | -0.139342 | 1 |
| 2 | -0.075200 | 1.908703 | -0.534952 | 0.042434 | -0.559363 | -0.252494 | -0.012058 | 0.284125 | 0 |
| 3 | -1.477081 | 0.865532 | -1.070530 | -0.451302 | 0.453224 | -0.201938 | -0.236852 | 0.128841 | 1 |
| 4 | -1.526756 | -0.336039 | 1.094536 | 0.286876 | -0.170061 | -0.175008 | -0.244818 | -0.050022 | 1 |
| 5 | -2.147808 | 0.475692 | -0.564722 | -0.344079 | -0.139551 | 0.425832 | -0.344405 | 0.070810 | 1 |
| 6 | 0.066967 | 1.802380 | -0.849193 | 0.543555 | -0.370788 | -0.232141 | 0.010738 | 0.268298 | 0 |
| 7 | -1.384515 | -0.041261 | 0.202822 | 1.299544 | 0.823427 | 0.123817 | -0.222009 | -0.006142 | 1 |
| 8 | 0.083638 | 0.888434 | 1.430529 | 0.522356 | 0.088537 | -0.931776 | 0.013412 | 0.132250 | 0 |
| 9 | -1.817628 | 0.741035 | 1.132660 | -0.646553 | 0.332754 | -0.019503 | -0.291460 | 0.110309 | 1 |
| 10 | -0.899886 | 0.776893 | -1.091563 | -0.339891 | 0.401541 | 0.382843 | -0.144298 | 0.115646 | 1 |
Now everything together
We will plot PC1 vs PC2 data. Each cluster will have a different color, and we will find the main feature for each principal component.
[24]:
plt.rcParams['axes.linewidth'] = 1.5
plt.figure(figsize=(10,8))
fig, ax = plt.subplots(figsize=(7,7))
color_code={ 0: 'magenta',\
1.0: 'orange',\
2.0: 'cyan',\
3.0: 'c',\
4.0: 'm',\
5.0: 'y',\
6.0: 'darkorange',
7.0: 'k',
}
for i in descriptors_pca.index:
ax.plot(descriptors_pca.loc[i].at['PC1_normalized'],descriptors_pca.loc[i].at['PC2_normalized'],
c=color_code[descriptors_pca.loc[i].at['Cluster_PC1_PC2']],
marker='o',markersize=8,markeredgecolor='k',alpha=0.3)
plt.xlabel ('PC1',fontsize=14,fontweight='bold')
ax.xaxis.set_label_coords(0.98, 0.45)
plt.ylabel ('PC2',fontsize=14,fontweight='bold')
ax.yaxis.set_label_coords(0.45, 0.98)
plt.tick_params ('both',width=2,labelsize=12)
ax.spines['left'].set_position(('data', 0))
ax.spines['bottom'].set_position(('data', 0))
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
lab=['MolWt', 'TPSA', 'nRotB', 'HBD','HBA', 'LogP'] #Feature labels
l=np.transpose(pca.components_[0:2, :]) ## We will get the components eigenvectors (main features) for PC1 and PC2
n = l.shape[0]
for i in range(n):
plt.arrow(0, 0, l[i,0], l[i,1],color= 'k',alpha=0.6,linewidth=1.2,head_width=0.025)
plt.text(l[i,0]*1.25, l[i,1]*1.25, lab[i], color = 'k',va = 'center', ha = 'center',fontsize=11)
circle = plt.Circle((0,0), 1, color='gray', fill=False,clip_on=True,linewidth=1.5,linestyle='--')
ax.add_artist(circle)
plt.xlim(-1.2,1.2)
plt.ylim(-1.2,1.2)
plt.tight_layout()
plt.show()
<Figure size 720x576 with 0 Axes>
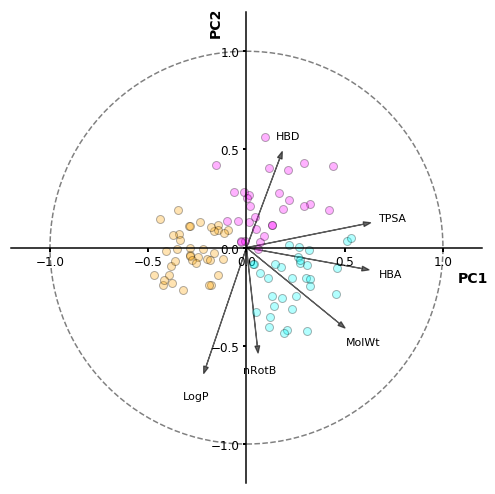
As a result, we can identify the features that correlate positively and negatively with PC1 (TPSA, HBA, MolWt), and with PC2 (HBD, nRotB, LogP). Additionally, we can see that LogP is the “most important” feature (descriptor) because of the vector length. And also we can see the 3 different clusters we identified by the silhouette-based algorithm.
Finally, we can merge our tables to keep the data in a single table or file.
[21]:
table=table.join(descriptors_pca)
[22]:
table.head(10)
[22]:
| SMILES | Id | MolWt | TPSA | nRotB | HBD | HBA | LogP | PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC1_normalized | PC2_normalized | Cluster_PC1_PC2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CC(C)C(C)(C)SCC=CBr | PV-002312932038 | 236.023434 | 0.00 | 4.0 | 0.0 | 1.0 | 4.0628 | -2.463784 | -0.936078 | -0.274030 | 0.558761 | -0.063431 | 0.131835 | -0.395072 | -0.139342 | 1 |
| 2 | CC1CC2(NC(C3CC3C)=NO2)C(C)O1 | Z3092470692 | 210.136828 | 42.85 | 1.0 | 1.0 | 4.0 | 1.4693 | -0.075200 | 1.908703 | -0.534952 | 0.042434 | -0.559363 | -0.252494 | -0.012058 | 0.284125 | 0 |
| 3 | BrC=CCN1CC2COC(C2)C1 | Z3310927277 | 231.025876 | 12.47 | 2.0 | 0.0 | 2.0 | 1.6157 | -1.477081 | 0.865532 | -1.070530 | -0.451302 | 0.453224 | -0.201938 | -0.236852 | 0.128841 | 1 |
| 4 | CC(C)CC(C)C(C)NCC1(C)CSC1 | Z3362561558 | 229.186421 | 12.03 | 6.0 | 1.0 | 2.0 | 3.3998 | -1.526756 | -0.336039 | 1.094536 | 0.286876 | -0.170061 | -0.175008 | -0.244818 | -0.050022 | 1 |
| 5 | CC(C)(C)CS(=O)CC=CCl | Z2904173473 | 194.053214 | 17.07 | 3.0 | 0.0 | 1.0 | 2.5337 | -2.147808 | 0.475692 | -0.564722 | -0.344079 | -0.139551 | 0.425832 | -0.344405 | 0.070810 | 1 |
| 6 | CC(C)(C)C1=NOC2(N1)[C@H]1OC[C@H]3C[C@@H]2C[C@@... | Z3092410916 | 236.152478 | 42.85 | 0.0 | 1.0 | 4.0 | 1.7169 | 0.066967 | 1.802380 | -0.849193 | 0.543555 | -0.370788 | -0.232141 | 0.010738 | 0.268298 | 0 |
| 7 | BrC=CCNC1CCCC=C1Br | Z3322438014 | 292.941474 | 12.03 | 3.0 | 1.0 | 1.0 | 3.3159 | -1.384515 | -0.041261 | 0.202822 | 1.299544 | 0.823427 | 0.123817 | -0.222009 | -0.006142 | 1 |
| 8 | CC(C)C1CC(NCCC2SCCS2)CN1 | Z3350500536 | 260.138091 | 24.06 | 5.0 | 2.0 | 4.0 | 2.1587 | 0.083638 | 0.888434 | 1.430529 | 0.522356 | 0.088537 | -0.931776 | 0.013412 | 0.132250 | 0 |
| 9 | FCC(CF)NCC1(C(F)F)CC1 | PV-002106209714 | 199.098412 | 12.03 | 6.0 | 1.0 | 1.0 | 1.9289 | -1.817628 | 0.741035 | 1.132660 | -0.646553 | 0.332754 | -0.019503 | -0.291460 | 0.110309 | 1 |
| 10 | CC(C(F)F)N1N=C(C(F)(F)F)C(F)C1=O | Z2790107381 | 248.038432 | 32.67 | 2.0 | 0.0 | 2.0 | 1.7386 | -0.899886 | 0.776893 | -1.091563 | -0.339891 | 0.401541 | 0.382843 | -0.144298 | 0.115646 | 1 |
Saving values from a pandas table to a .csv file is very easy. You just need to type:
table.to_csv('file_name.csv')